Did you know that there is a nest of endangered long nosed bandicoots living just beside the popular Manly beach in Sydney, Australia? Well, I didn't, until I looked at BioNet. The Australian NSW government created BioNet as a government database of all flora and fauna species sightings in NSW. It's absolutely fantastic. If you're an architect and want to see how you might impact the urban ecosystem in NSW, look at BioNet. If you're an ecologist of some kind, you probably already use it. If you're just a good citizen who wants to remodel your back yard to improve urban ecology, BioNet is there for you.
Fortunately, BioNet comes with an online search system called Atlas. It's simple to use, but unfortunately it has limits on the data it produces. It won't show you all the fields associated with species, won't show meta fields, and has a limit to the quantity of records shown. Thankfully, BioNet comes with an API which can be queried with programming knowledge. I've written a bit of Python which will allow you to download regions of data; but before we get to that, let's see a graphic!
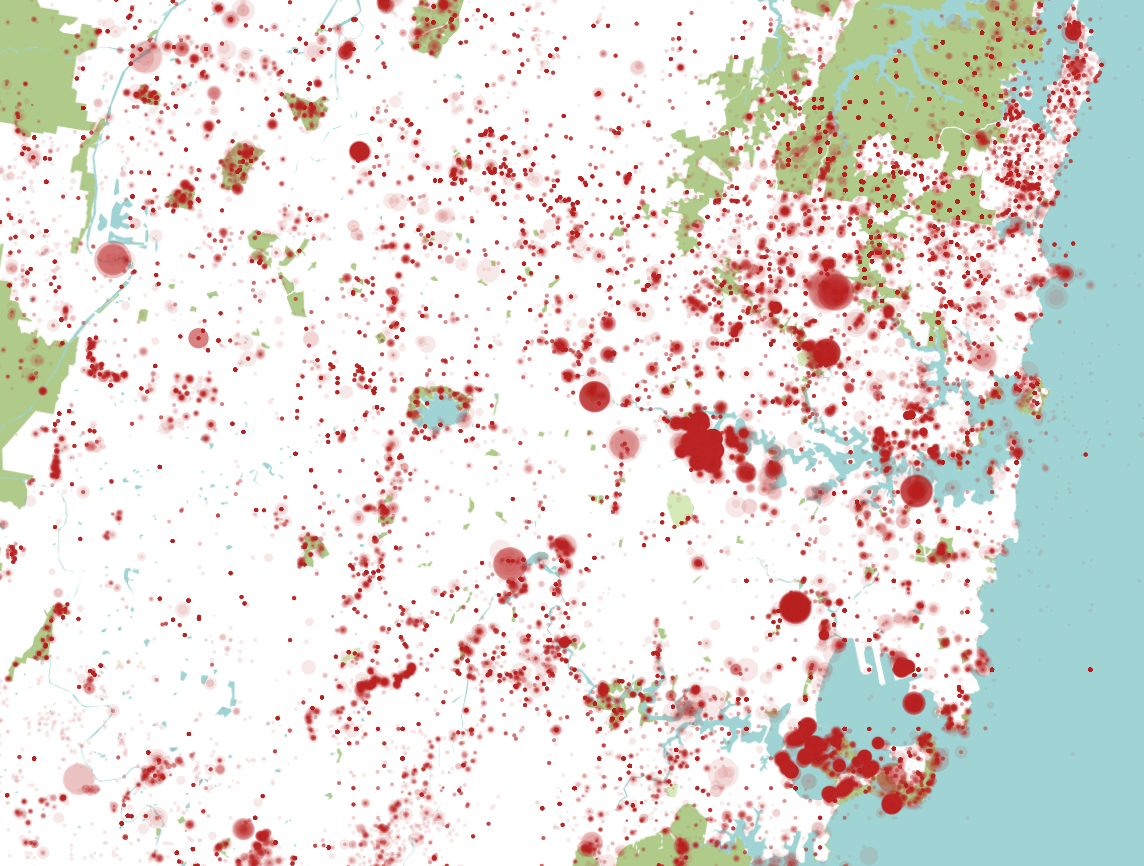
I've plotted every species on the database close to Sydney in the map above. Size is relative to the number of species sighted (logarithmic relationship). I haven't done any real filtering beyond this, so it's not very meaningful, but it shows the data and shows it can be geolocated. It also looks like someone murdered the country, but I'll post the interesting visualisations in a future post.
The Python code works in two parts. The first queries the API for json
results divided into square tiles from a top left and bottom right
latitude and longitude coordinate region. This'll give you a bunch of
*.json files in the current working directory. Edit the coordinates
and resolution as necessary, and off you go. I've put in a series of
fields that should be good for more general uses, but you can check the
BioNet Data API for all fields.
import os
start = (-33.408554, 150.326152)
end = (-34.207799, 151.408916)
lat = start[0]
lon = start[1]
def create_url(lat, lon, lat_next, lon_next):
return 'https://data.bionet.nsw.gov.au/biosvcapp/odata/SpeciesSightings_CoreData?$select=kingdom,catalogNumber,basisOfRecord,dcterms_bibliographicCitation,dataGeneralizations,informationWithheld,dcterms_modified,dcterms_available,dcterms_rightsHolder,IBRASubregion,scientificName,vernacularName,countryConservation,stateConservation,protectedInNSW,sensitivityClass,eventDate,individualCount,observationType,status,coordinateUncertaintyInMeters,decimalLatitude,decimalLongitude,geodeticDatum&$filter=((decimalLongitude ge ' + str(lon) + ') and (decimalLongitude le ' + str(lon_next) + ')) and ((decimalLatitude le ' + str(lat) + ') and (decimalLatitude ge ' + str(lat_next) + '))'
i = 0
resolution = 0.05
while (lat > end[0]):
while (lon < end[1]):
lat_next = round(lat - resolution, 6)
lon_next = round(lon + resolution, 6)
url = create_url(lat, lon, lat_next, lon_next).replace(' ', '%20').replace('\'', '%27')
os.system('curl \'' + url + "\' -H 'Host: data.bionet.nsw.gov.au' -H 'User-Agent: Mozilla/5.0 (X11; Linux x86_64; rv:52.0) Gecko/20100101 Firefox/52.0' -H 'Accept: text/html,application/xhtml+xml,application/xml;q=0.9,*/*;q=0.8' -H 'Accept-Language: en-US,en;q=0.5' --compressed -H 'Cookie: NSC_EBUB_CJPOFU_443_mcwjq=ffffffff8efb154f45525d5f4f58455e445a4a423660' -H 'DNT: 1' -H 'Connection: keep-alive' -H 'Upgrade-Insecure-Requests: 1' -H 'Cache-Control: max-age=0' > " + str(i) + '.json')
i += 1
lon = round(lon + resolution, 6)
lon = start[1]
lat = round(lat - resolution, 6)
Now we'll run another little script which will convert all the json
files in the directory into a single csv file. You can read this csv
file in programs like Excel or QGIS for further analysis.
import unicodecsv as csv
import json
f = csv.writer(open('bionet.csv', 'wb+'), encoding='utf-8')
number_of_json_files = 352
f.writerow([
'IBRASubregion',
'basisOfRecord',
'catalogNumber',
'coordinateUncertaintyInMeters',
'countryConservation',
'dataGeneralizations',
'dcterms_available',
'dcterms_bibliographicCitation',
'dcterms_modified',
'dcterms_rightsHolder',
'decimalLatitude',
'decimalLongitude',
'eventDate',
'geodeticDatum',
'individualCount',
'informationWithheld',
'observationType',
'protectedInNSW',
'scientificName',
'sensitivityClass',
'stateConservation',
'status',
'kingdom',
'vernacularName',
])
i = 0
while i < number_of_json_files:
data = json.load(open(str(i) + '.json'))
print(i)
for x in data['value']:
f.writerow([
x['IBRASubregion'],
x['basisOfRecord'],
x['catalogNumber'],
x['coordinateUncertaintyInMeters'],
x['countryConservation'],
x['dataGeneralizations'],
x['dcterms_available'],
x['dcterms_bibliographicCitation'],
x['dcterms_modified'],
x['dcterms_rightsHolder'],
x['decimalLatitude'],
x['decimalLongitude'],
x['eventDate'],
x['geodeticDatum'],
x['individualCount'],
x['informationWithheld'],
x['observationType'],
x['protectedInNSW'],
x['scientificName'],
x['sensitivityClass'],
x['stateConservation'],
x['status'],
x['kingdom'],
x['vernacularName'],
])
i += 1
That's it! Have fun and don't forget to check for frogs in your backyards. If you don't have any, build a pond. Or at least a water bath for the birds.